This chapter examines and imputes missing observations in the gridded weather data from Chapter 3 .
── Attaching core tidyverse packages ──────────────────────── tidyverse 2.0.0 ──
✔ dplyr 1.1.4 ✔ readr 2.1.5
✔ forcats 1.0.0 ✔ stringr 1.5.2
✔ ggplot2 4.0.0 ✔ tibble 3.3.0
✔ lubridate 1.9.4 ✔ tidyr 1.3.1
✔ purrr 1.1.0
── Conflicts ────────────────────────────────────────── tidyverse_conflicts() ──
✖ dplyr::filter() masks stats::filter()
✖ dplyr::lag() masks stats::lag()
ℹ Use the conflicted package (<http://conflicted.r-lib.org/>) to force all conflicts to become errors
Linking to GEOS 3.13.0, GDAL 3.8.5, PROJ 9.5.1; sf_use_s2() is TRUE
Let us load the graphs theme functions (See Chapter 1 ):
source ("../scripts/functions/utils.R" )
Let us also load the maps for the countries of interest (See Chapter 2 ).
load ("../data/Maps/GADM-4.1/maps_level_0.RData" )<- st_shift_longitude (maps_level_0$ NZL)
We focus on the countries defined in Chapter 2 :
load ("../output/tb_countries.rda" )
# A tibble: 1 × 4
country_code country_name country_name_short country_name_clear
<chr> <chr> <chr> <chr>
1 NZL New Zealand New Zealand New-Zealand
Import Data
We define a function, load_daily ()Chapter 3 .
#' Loads a weather data from country x year #' #' @param file path to the file to load <- function (file) {load (file)
Let us focus on New Zealand.
We extract the corresponding country code:
<- tb_countries |> :: filter (country_name == !! country) |> :: pull ("country_code" )
We list the files that contain the daily data for the current country:
<- list (precip = "../Data/Weather/CPC/grid_daily_precip/" ,tmin = "../Data/Weather/CPC/grid_daily_tmin/" ,tmax = "../Data/Weather/CPC/grid_daily_tmax/" |> map (~ list.files (.x, full.names = TRUE , pattern = str_c (country_code, "_" )))
Let us load these files:
# Load precip, tmin and tmax <- map (N$ precip, load_daily) |> list_rbind ()<- map (N$ tmin, load_daily) |> list_rbind ()<- map (N$ tmax, load_daily) |> list_rbind ()
Let us merge these datasets:
<- country_precip_daily |> :: mutate (variable = "precip" ) |> bind_rows (|> mutate (variable = "tmin" )|> bind_rows (|> mutate (variable = "tmax" )|> :: select (- grid_id) |> pivot_wider (values_from = value, names_from = variable) |> group_by (longitude, latitude) |> :: mutate (cell_id = cur_group_id ()) |> ungroup ()
We extract information about longitude / latitude and geometry of each cell:
<- country_weather_daily |> :: select (cell_id, longitude, latitude, geometry, intersect_area) |> unique ()<- cells |> dplyr:: mutate (country = !! country)
Let us transform this object as an sf object:
<- st_as_sf (cells)
We can have a look at the number of values per cell.
# This code is not evaluated here. #| code-fold: true ggplot () + geom_sf (data = st_as_sf (cells) |> left_join (|> group_by (cell_id) |> count () |> arrange (n),by = "cell_id" mapping = aes (fill = n)+ geom_sf (data = maps_level_0_NZ$ NGA,fill = NA , col = "black"
We can identify the cells for which the number of records is lower than the max number of records:
<- |> group_by (cell_id) |> count () |> ungroup () |> mutate (max = max (n)) |> filter (n < max)
# This code is not evaluated here. #| code-fold: true ggplot () + geom_sf (data = st_as_sf (cells) |> left_join (|> group_by (cell_id) |> count () |> arrange (n),by = "cell_id" |> left_join (|> select (cell_id) |> mutate (to_remove = TRUE )|> mutate (to_remove = replace_na (to_remove, FALSE )),mapping = aes (fill = to_remove)+ geom_sf (data = maps_level_0_NZ,fill = NA , col = "black" + scale_fill_manual ("Remove" , values = c (` TRUE ` = "red" , ` FALSE ` = NA )
Let us remove those cells:
<- |> filter (! cell_id %in% cells_to_remove$ cell_id)
We also remove the cells corresponding to islands for which we will have no agricultural records.
<- c (1 , 2 , 5 , 6 , 24 , 25 , 34 , 35 , 45 , 46 , 55 , 56 , 256 , 250 , 249 , 255 ,264 , 267 , 263 , 266 , 261 , 260 , 259 , 262 , 265 <- |> filter (! cell_id %in% ids_islands
Code
# This chunk is not evaluated here. ggplot () + geom_sf (data = st_as_sf (cells) |> inner_join (|> group_by (cell_id) |> count () |> arrange (n),by = "cell_id" fill = "red" + geom_sf (data = maps_level_0_NZ,fill = NA , col = "black"
<- |> filter (! cell_id %in% c (cells_to_remove$ cell_id, ids_islands)<- st_as_sf (cells)
Let us save this grid:
save (file = str_c ("../data/Weather/CPC/" , country_code, "_cells.RData" )
We remove the geometry from the table:
<- |> :: select (- geometry, - intersect_area)
The total number of observations:
:: number (nrow (country_weather_daily), big.mark = "," )
Missing Values
How many missing values are there here in the dataset?
|> summarise (across (everything (), ~ sum (is.na (.x))))
# A tibble: 1 × 8
longitude latitude country_code date precip tmin tmax cell_id
<int> <int> <int> <int> <int> <int> <int> <int>
1 0 0 0 0 16895 446893 445241 0
Precipitation
The number of missing data for precipitation per cell:
<- country_weather_daily |> group_by (cell_id) |> select (cell_id, precip) |> summarise (nb_na_precip = sum (is.na (precip))) |> arrange (desc (nb_na_precip))
# A tibble: 181 × 2
cell_id nb_na_precip
<int> <int>
1 153 9875
2 9 39
3 12 39
4 13 39
5 14 39
6 19 39
7 20 39
8 21 39
9 22 39
10 27 39
# ℹ 171 more rows
What if we look at missing values per cell and day?
|> group_by (date) |> summarise (nb_na_precip = sum (is.na (precip))) |> filter (nb_na_precip > 0 ) |> arrange (desc (nb_na_precip)) |> :: datatable ()
And by cell?
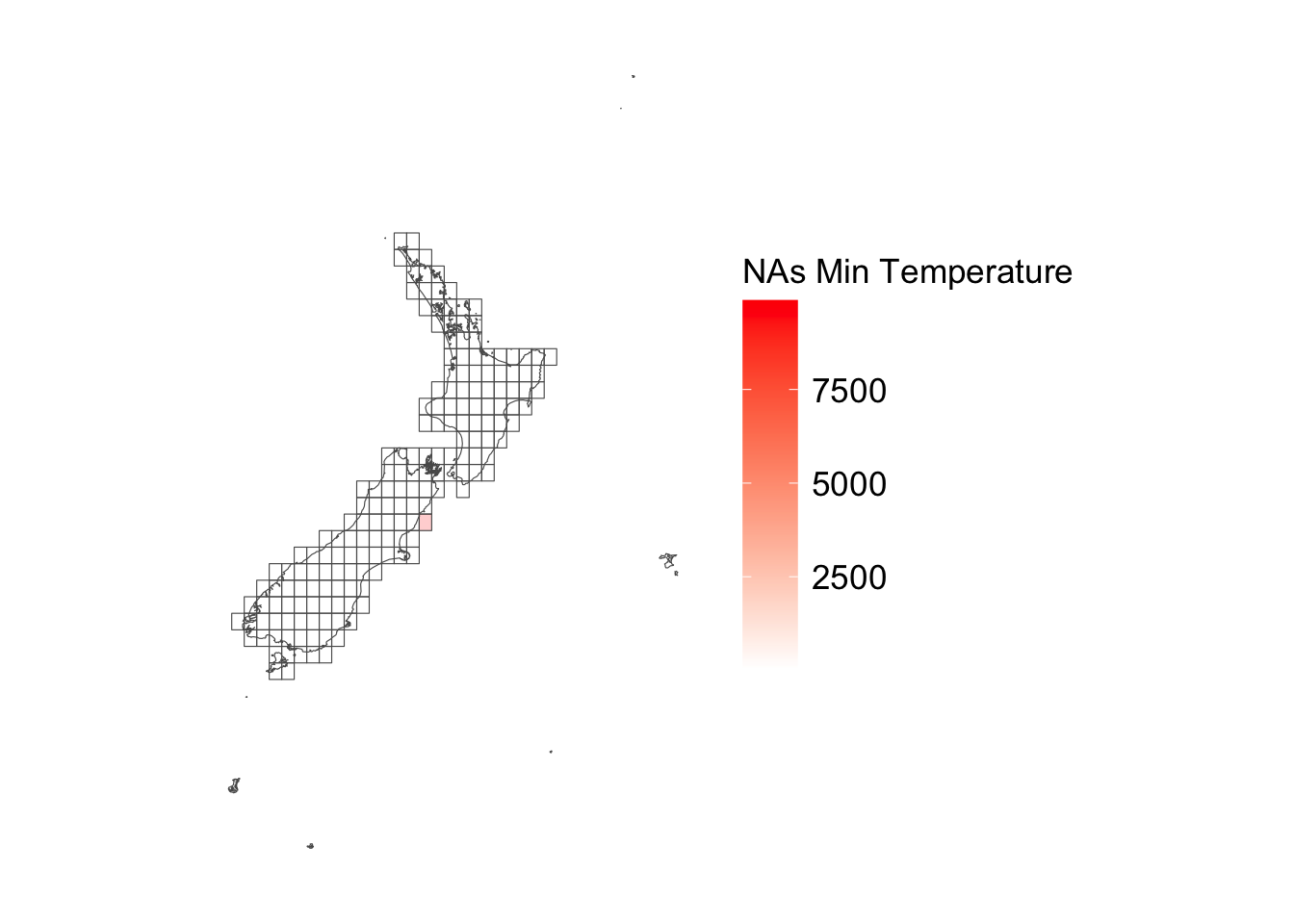
ggplot () + geom_sf (data = cells_sf |> left_join (tb_nb_na_precip),alpha = .2 ,mapping = aes (fill = nb_na_precip)+ geom_sf (data = maps_level_0_NZ, fill = NA ) + scale_fill_gradient ("NAs Min Temperature" , low = "white" , high = "red" ) + theme_map_paper ()
Joining with `by = join_by(cell_id)`
Most missing precipitation observations are from cell 153 which is covering a tiny fraction of land.
Minimum Temperatures
Now, let us do the same for min temperatures:
<- country_weather_daily |> group_by (cell_id) |> select (cell_id, tmin) |> summarise (nb_na_tmin = sum (is.na (tmin))) |> arrange (desc (nb_na_tmin))
# A tibble: 181 × 2
cell_id nb_na_tmin
<int> <int>
1 152 16437
2 171 16437
3 196 16437
4 221 16437
5 239 16437
6 258 16437
7 9 6979
8 14 6979
9 22 6979
10 27 6979
# ℹ 171 more rows
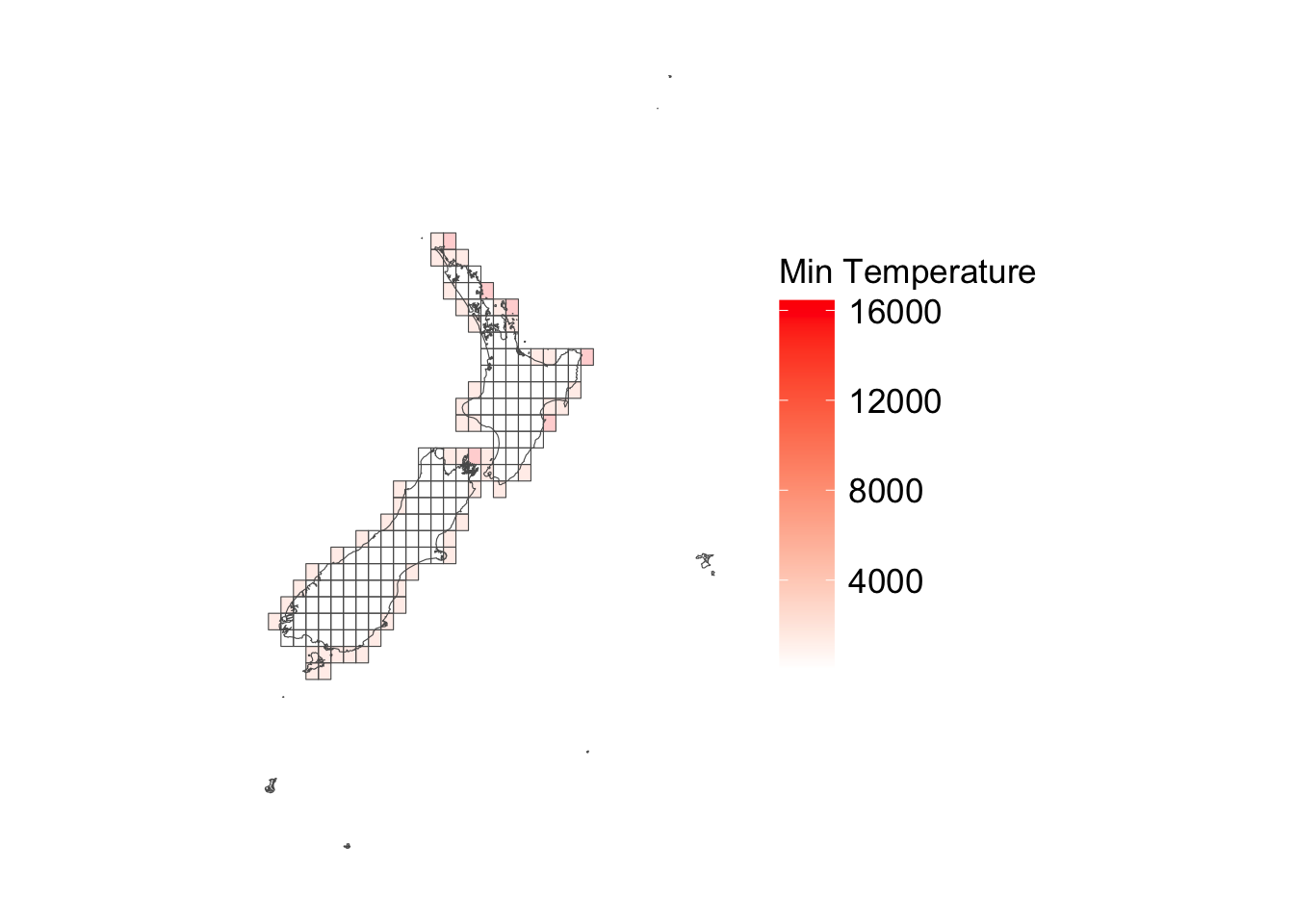
ggplot () + geom_sf (data = cells_sf |> left_join (tb_nb_na_tmin),alpha = .2 ,mapping = aes (fill = nb_na_tmin)+ geom_sf (data = maps_level_0_NZ, fill = NA ) + scale_fill_gradient ("Min Temperature" , low = "white" , high = "red" ) + theme_map_paper ()
Joining with `by = join_by(cell_id)`
Spatially, missing observation are mostly on coastal areas.
For each cell, the number of daily observations from 1980-01-01 to 2024-12-31 is:
seq (min (country_weather_daily$ date), max (country_weather_daily$ date), by = "day" |> length ()
What if we look at missing values per cell and day?
|> group_by (date) |> summarise (nb_na_tmin = sum (is.na (tmin))) |> filter (nb_na_tmin > 10 ) |> arrange (desc (nb_na_tmin)) |> :: datatable ()
Maximum Temperatures
And for max temperatures:
<- |> group_by (cell_id) |> select (cell_id, tmax) |> summarise (nb_na_tmax = sum (is.na (tmax))) |> arrange (desc (nb_na_tmax))
# A tibble: 181 × 2
cell_id nb_na_tmax
<int> <int>
1 152 16437
2 171 16437
3 196 16437
4 221 16437
5 239 16437
6 258 16437
7 9 6971
8 14 6971
9 22 6971
10 27 6971
# ℹ 171 more rows
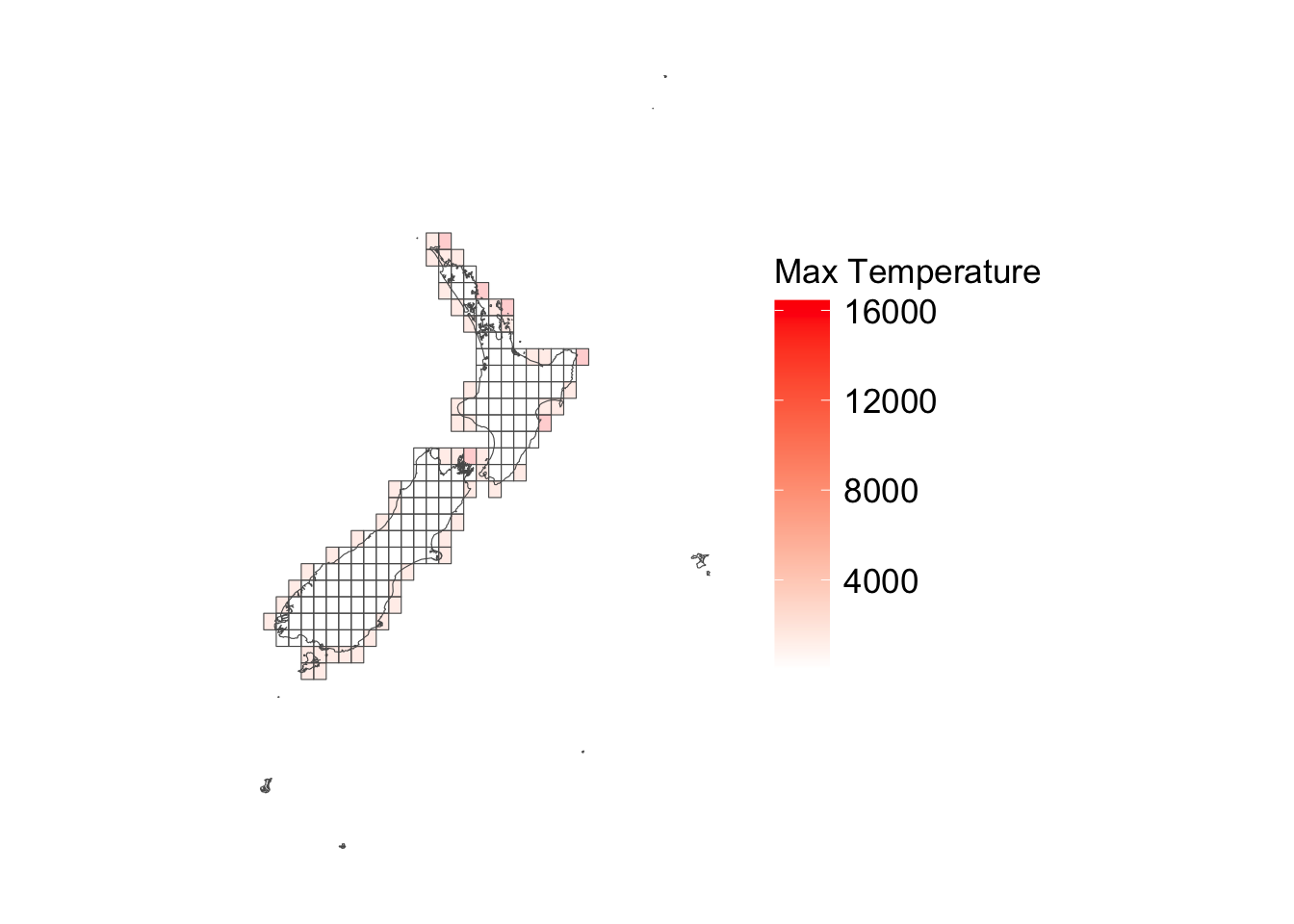
ggplot () + geom_sf (data = cells_sf |> left_join (tb_nb_na_tmax),alpha = .2 ,mapping = aes (fill = nb_na_tmax)+ geom_sf (data = maps_level_0_NZ, fill = NA ) + scale_fill_gradient ("Max Temperature" , low = "white" , high = "red" ) + theme_map_paper ()
Joining with `by = join_by(cell_id)`
What if we look at missing values per cell and day?
|> group_by (date) |> summarise (nb_na_tmax = sum (is.na (tmax))) |> filter (nb_na_tmax > 10 ) |> arrange (desc (nb_na_tmax)) |> :: datatable ()
Temporal Interpolation
Let us use splines at the cell level to interpolate missing values for which no more than 5 consecutive values are missing. We will then address the interpolation of missing values for the south-western cells.
<- |> arrange (date) |> nest (.by = cell_id) |> mutate (precip_new = map (data, ~ {if (sum (is.na (.x$ precip)) >= nrow (.x)- 1 ) {<- NA else {<- imputeTS:: na_interpolation (.x$ precip, maxgap = 5 , option = "spline" )tmin_new = map (data, ~ {if (sum (is.na (.x$ tmin)) >= nrow (.x)- 1 ) {<- NA else {<- imputeTS:: na_interpolation (.x$ tmin, maxgap = 5 , option = "spline" )tmax_new = map (data, ~ {if (sum (is.na (.x$ tmax)) >= nrow (.x)- 1 ) {<- NA else {<- imputeTS:: na_interpolation (.x$ tmax, maxgap = 5 , option = "spline" )|> unnest (cols = c (data, precip_new, tmin_new, tmax_new))
Registered S3 method overwritten by 'quantmod':
method from
as.zoo.data.frame zoo
With the interpolation, there might be negative values, which is not possible for precipitation. We set to 0 precipitation for which interpolated values are negative.
<- |> mutate (precip_new = ifelse (precip_new < 0 , 0 , precip_new)
Spatial Interpolation
Now, let us address the problem of the remaining cells for which we have some missing observation. First, we identify cells IDs for which observations are missing for precipitation, and for min and max temperatures.
<- |> pull (cell_id)<- |> pull (cell_id)<- |> pull (cell_id)
We will replace the missing observations with weighted averages of the values found in the neighbors cells. We will consider the 5 nearest cells and use the inverse distance to those as the weights in the weighted average.
First, let us get the distance matrix of the cells, using the centroid of each cell as the reference point.
<- sf:: st_centroid (cells_sf)
Warning: st_centroid assumes attributes are constant over geometries
<- sf:: st_distance (centroids_cells, centroids_cells)
We can then define the find_neighbors_radius ()
#' Identifies the neighbors of a cell given a radius #' @param id cell id #' @param radius radius in metres to identify the neighbors <- function (id, radius) {<- which (cells_sf$ cell_id == id)tibble (cell_id = id,cell_id_neighbors = cells_sf$ cell_id,distance = units:: drop_units (distance_matrix[, ind_current_cell])|> arrange (distance) |> filter (cell_id_neighbors != !! id) |> filter (distance <= !! radius)
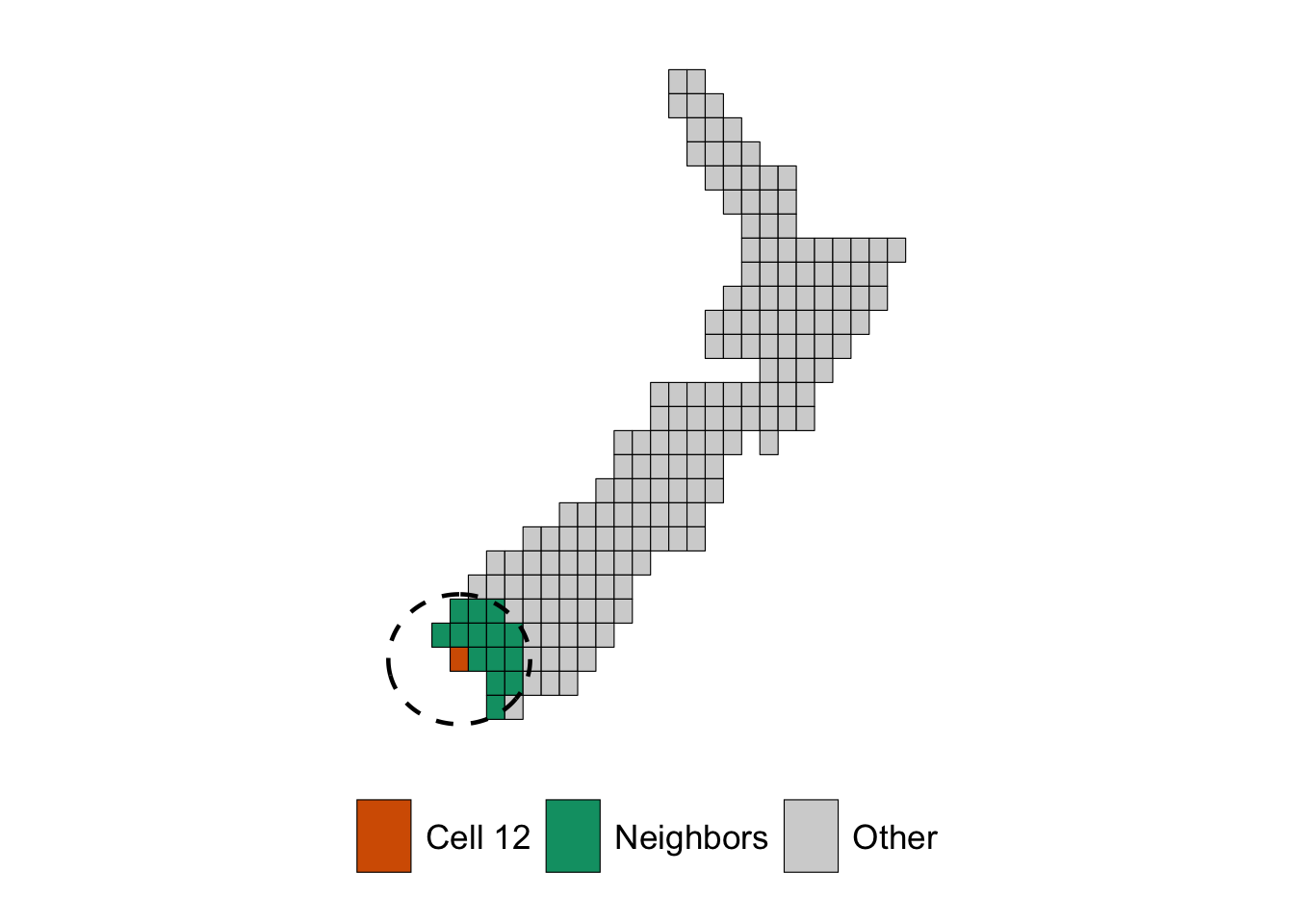
For example, for cell ID 12, the cells within 150km radius:
<- find_neighbors_radius (id = 12 , 150 * 1000 )
# A tibble: 14 × 3
cell_id cell_id_neighbors distance
<dbl> <int> <dbl>
1 12 19 38446.
2 12 13 55597.
3 12 9 67695.
4 12 20 67695.
5 12 29 76893.
6 12 28 94601.
7 12 30 95169.
8 12 14 111194.
9 12 39 115338.
10 12 21 117767.
11 12 38 127563.
12 12 40 128510.
13 12 27 134789.
14 12 31 135586.
Code
<- function (long, lat, radius){ tibble (degree = 1 : 360 ) |> rowwise () |> mutate (long = geosphere:: destPoint (c (long, lat), degree, radius)[1 ],lat = geosphere:: destPoint (c (long, lat), degree, radius)[2 ]|> ungroup ()<- centroids_cells |> filter (cell_id == 12 )<- create_circle (long = cell_of_interest$ longitude,lat = cell_of_interest$ latitude,radius = 150 * 1000 |> st_as_sf (coords = c ("long" , "lat" ), crs = sf:: st_crs (centroids_cells)) |> summarise (geometry = st_combine (geometry)) %>% st_cast ("POLYGON" )ggplot () + geom_sf (data = cells_sf |> mutate (type = case_when (== 12 ~ "Cell 12" ,%in% example_nn$ cell_id_neighbors ~ "Neighbors" ,TRUE ~ "Other" mapping = aes (fill = type),colour = "black" + geom_sf (data = maps_level_0$ NGA, fill = NA ) + geom_sf (data = circle_sf, color = "black" , fill = NA , linewidth = .8 , linetype = "dashed" + scale_fill_manual (NULL , values = c ("Cell 12" = "#D55E00" , "Neighbors" = "#009E73" , "Other" = "lightgray" )+ theme_map_paper () + theme (legend.position = "bottom" )
Now, let us get all the neighbors of the cells with many missing values for min temperatures, and for max temperatures.
<- map (cell_id_missing_precip, ~ find_neighbors_radius (id = .x, radius = 150 * 1000 )) |> list_rbind ()<- map (cell_id_missing_tmin, ~ find_neighbors_radius (id = .x, radius = 150 * 1000 )) |> list_rbind ()<- map (cell_id_missing_tmax, ~ find_neighbors_radius (id = .x, radius = 150 * 1000 )) |> list_rbind ()
We then define the impute_na_neighbor_cells ()name_col) from the neighbor (found in argument neighbors) cells of a given cell (whose ID is given to the argument id).
#' @param id cell ID #' @param neighbors tibble with distances of the neighbors (obtained with #' `find_neighbors_radius()`) #' @param name_col name of the columns in `country_weather_daily` to use to #' impute data (from neighbors) <- function (id, neighbors, name_col) {|> filter (cell_id == !! id) |> left_join (neighbors, relationship = "many-to-many" , by = "cell_id" ) |> left_join (|> select (cell_id_neighbors = cell_id, new_val_neighbors = !! sym (name_col)|> :: st_drop_geometry (),by = c ("date" , "cell_id_neighbors" )|> select (date, cell_id, cell_id_neighbors, new_val_neighbors, distance) |> filter (! is.na (new_val_neighbors)) |> group_by (cell_id, date) |> mutate (weight_distance = distance / sum (distance)) |> mutate (new_val_neighbors = new_val_neighbors * weight_distance) |> summarise (new_val_neighbors = sum (new_val_neighbors),.groups = "drop"
We apply this function to the cells for which many missing values were found, to impute values for min temperatures, and then for max temperatures.
<- map (~ impute_na_neighbor_cells (neighbors = neighbors_precip, name_col = "precip_new" )|> list_rbind ()<- map (~ impute_na_neighbor_cells (neighbors = neighbors_tmin, name_col = "tmin_new" )|> list_rbind ()<- map (~ impute_na_neighbor_cells (neighbors = neighbors_tmax, name_col = "tmax_new" )|> list_rbind ()
We replace the missing values in the table with daily observations:
<- |> left_join (|> rename (precip_neighbors = new_val_neighbors)|> left_join (|> rename (tmin_neighbors = new_val_neighbors)|> left_join (|> rename (tmax_neighbors = new_val_neighbors)|> mutate (# If temporal interpolation was not enough, replace with spatially # interpolated values precip_new = ifelse (is.na (precip_new), precip_neighbors, precip_new),tmin_new = ifelse (is.na (tmin_new), tmin_neighbors, tmin_new),tmax_new = ifelse (is.na (tmax_new), tmax_neighbors, tmax_new)
Second Temporal Interpolation
There are still missing values:
precip: 2015-05-19 to 2015-05-26,tmin and tmax: 1983-04-25 to 1983-04-30 and 2015-05-21 to 2015-05-26.
|> filter (is.na (precip_new)) |> count (date)|> filter (is.na (tmin_new)) |> count (date)|> filter (is.na (tmax_new)) |> count (date)
For those observations, we use, again, a temporal interpolation. This time, we increase the windows’ width to 10 days.
<- |> arrange (date) |> nest (.by = cell_id) |> mutate (precip_new_2 = map (data, ~ {if (sum (is.na (.x$ precip_new)) >= nrow (.x)- 1 ) {<- NA else {<- imputeTS:: na_interpolation (.x$ precip_new, maxgap = 10 , option = "spline" )tmin_new_2 = map (data, ~ {if (sum (is.na (.x$ tmin_new)) >= nrow (.x)- 1 ) {<- NA else {<- imputeTS:: na_interpolation (.x$ tmin_new, maxgap = 10 , option = "spline" )tmax_new_2 = map (data, ~ {if (sum (is.na (.x$ tmax_new)) >= nrow (.x)- 1 ) {<- NA else {<- imputeTS:: na_interpolation (.x$ tmax_new, maxgap = 10 , option = "spline" )|> unnest (cols = c (data, precip_new_2, tmin_new_2, tmax_new_2)) |> mutate (precip_new_2 = ifelse (precip_new_2 < 0 , 0 , precip_new_2),precip_new = ifelse (is.na (precip_new), precip_new_2, precip_new),tmin_new = ifelse (is.na (tmin_new), tmin_new_2, tmin_new),tmax_new = ifelse (is.na (tmax_new), tmax_new_2, tmax_new)
Lastly, we remove the raw values with missing data with the columns in which missing data were replaced.
<- |> :: select (- precip, - tmin, - tmax, - precip_neighbors, - tmin_neighbors, - tmax_neighbors,- precip_new_2, - tmin_new_2, - tmax_new_2|> rename (precip = precip_new, tmin = tmin_new, tmax = tmax_new)
Let us save the results for later use.
save (file = "../data/Weather/CPC/NZL_daily_weather_corrected.rda"
Each row gives three metrics for a grid cell identified with its ID (cell_id) whose centroid’s coordinates are given in longitude and latitude, at a given day (date). The country the data refer to (Nigeria) is stated in column country_code. The metrics are total rainfall (precip), minimum temperature (tmin) and maximum temperature (tmax).
The content of this dataset is summarized in Table 4.1 .