Let us first load {tidyverse}:
── Attaching core tidyverse packages ──────────────────────── tidyverse 2.0.0 ──
✔ dplyr 1.1.4 ✔ readr 2.1.5
✔ forcats 1.0.0 ✔ stringr 1.5.1
✔ ggplot2 3.5.1 ✔ tibble 3.2.1
✔ lubridate 1.9.3 ✔ tidyr 1.3.1
✔ purrr 1.0.2
── Conflicts ────────────────────────────────────────── tidyverse_conflicts() ──
✖ dplyr::filter() masks stats::filter()
✖ dplyr::lag() masks stats::lag()
ℹ Use the conflicted package (<http://conflicted.r-lib.org/>) to force all conflicts to become errors
Download and extract the weather data
The data is available in monthly summaries. We need to create some function to download those summaries and then extract the information.
#' Downloads the monthly summary of the weather. #' Puts the results in `data/meteo/tmp/` #' #' @param lien url of the summary to be downloaded #' @return returns nothing #' lien <- paste0( #' "https://donneespubliques.meteofrance.fr/donnees_libres/", #' "Txt/Climat/climat.201903.csv.gz" #' ) <- function (lien) {<- |> str_split ("/" ) |> last () |> last ()download.file (lien, destfile = str_c ("../data/meteo/tmp/" , filename))
Let us loop over the months from 1990 to 2019. At each iteration, we can download the monthy summary:
<- txtProgressBar (min = 1990 , max = 2019 , style = 2 )for (year in 1990 : 2019 ) {for (month in 1 : 12 ) {<- str_c ("https://donneespubliques.meteofrance.fr/" ,"donnees_libres/Txt/Climat/" ,"climat." , year,str_pad (month, width = 2 , side = "left" , pad = 0 ), ".csv.gz" telecharger_mess (lien)setTxtProgressBar (pb = pb, year)
We need to get some information relative to each weather station. The list of weather stations is available online . We have downloaded it manually and put it in data/meteo/.
<- read_delim ("../data/meteo/postesSynop.csv" , locale = locale (decimal_mark = "." ),delim = ";"
Rows: 62 Columns: 5
── Column specification ────────────────────────────────────────────────────────
Delimiter: ";"
chr (2): ID, Nom
dbl (3): Latitude, Longitude, Altitude
ℹ Use `spec()` to retrieve the full column specification for this data.
ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.
# A tibble: 62 × 5
ID Nom Latitude Longitude Altitude
<chr> <chr> <dbl> <dbl> <dbl>
1 07005 ABBEVILLE 50.1 1.83 69
2 07015 LILLE-LESQUIN 50.6 3.10 47
3 07020 PTE DE LA HAGUE 49.7 -1.94 6
4 07027 CAEN-CARPIQUET 49.2 -0.456 67
5 07037 ROUEN-BOOS 49.4 1.18 151
6 07072 REIMS-PRUNAY 49.2 4.16 95
7 07110 BREST-GUIPAVAS 48.4 -4.41 94
8 07117 PLOUMANAC'H 48.8 -3.47 55
9 07130 RENNES-ST JACQUES 48.1 -1.73 36
10 07139 ALENCON 48.4 0.110 143
# ℹ 52 more rows
Let us list all the downloaded monthly summaries:
<- list.files ("../data/meteo/tmp" , pattern = " \\ .csv \\ .gz" , full.names = TRUE
We can extract the data from all these files. To that end, let us create a function that will be applied to all these files.
#' Extracts sunlight data from a monthly weather summary #' #' @param fichier path to the file to extract data from #' fichier <- N[1] <- function (fichier) {<- read_csv2 (gzfile (fichier))gzfile (fichier) |> read_delim (locale = locale (decimal_mark = "." ), delim = ";" ) |> select (NUM_POSTE, DAT, INST)
We extract the following columns:
NUM_POSTE: OMM ID of the weather stationDAT: date of observation (year and month)INST: cumulated sunlight data (in number of hours)
Let us loop over the downloaded files:
<- map_df (N, extract_data)
We only keep stations where at least 200 records are available:
<- |> group_by (NUM_POSTE) |> tally () |> arrange (n) |> filter (n > 200 )<- |> filter (NUM_POSTE %in% stations_to_keep$ NUM_POSTE)
Leaving us with:
Let us compute the monthly average:
<- |> mutate (month = lubridate:: month (DAT)) |> group_by (NUM_POSTE, month) |> filter (DAT < lubridate:: ymd ("2011-12-31" )) |> summarise (INST = mean (INST, na.rm= TRUE ))
`summarise()` has grouped output by 'NUM_POSTE'. You can override using the
`.groups` argument.
And let us add the stations informations:
<- |> left_join (stations, by = c ("NUM_POSTE" = "ID" ))
# A tibble: 684 × 7
# Groups: NUM_POSTE [57]
NUM_POSTE month INST Nom Latitude Longitude Altitude
<chr> <dbl> <dbl> <chr> <dbl> <dbl> <dbl>
1 07005 1 68.4 ABBEVILLE 50.1 1.83 69
2 07005 2 78.2 ABBEVILLE 50.1 1.83 69
3 07005 3 128. ABBEVILLE 50.1 1.83 69
4 07005 4 181. ABBEVILLE 50.1 1.83 69
5 07005 5 205 ABBEVILLE 50.1 1.83 69
6 07005 6 207 ABBEVILLE 50.1 1.83 69
7 07005 7 220. ABBEVILLE 50.1 1.83 69
8 07005 8 208. ABBEVILLE 50.1 1.83 69
9 07005 9 159. ABBEVILLE 50.1 1.83 69
10 07005 10 114 ABBEVILLE 50.1 1.83 69
# ℹ 674 more rows
Interpolation of sunlight data: Kriging
We will perform some kriging methods to interpolate the average monthly sunlight to each point of a grid covering Metropolitan France. We load a shapefile of France (from Institut National de l’Information Géographique et Forestière). The data can be downloaded from the French Open platform for French public data: Geofla .
At the level of French “départements”
Linking to GEOS 3.11.0, GDAL 3.5.3, PROJ 9.1.0; sf_use_s2() is TRUE
<- :: read_sf ("../data/France_departements_ign/DEPARTEMENT.SHP" ) |> :: st_transform (crs = 4326 )
Let us extract a bounding box for France:
<- st_bbox (map_departements)
xmin ymin xmax ymax
-5.138996 41.362165 9.559615 51.089001
Let us keep only weather stations located in Metropolitan France:
<- |> filter (>= bbox_france["xmin" ],<= bbox_france["xmax" ],>= bbox_france["ymin" ],<= bbox_france["ymax" ]
The coordinates of these weather stations:
<- |> select ("Longitude" , "Latitude" ) |> unique () |> st_as_sf (coords = c ("Longitude" , "Latitude" )) |> :: st_set_crs (4326 )
Adding missing grouping variables: `NUM_POSTE`
Let us now define the grid (thanks to Jareck Kotowski for answering his own question on StackOverflow ):
<- .5 <- seq (bbox_france["xmin" ], bbox_france["xmax" ], by = size_cell)<- seq (bbox_france["ymin" ], bbox_france["ymax" ], by = size_cell)<- expand.grid (long = long_grid, lat = lat_grid)head (france_grid)
long lat
1 -5.138996 41.36216
2 -4.638996 41.36216
3 -4.138996 41.36216
4 -3.638996 41.36216
5 -3.138996 41.36216
6 -2.638996 41.36216
We need to change this grid to a sf object:
<- |> st_as_sf (coords = c ("long" , "lat" ), crs = 4326 , agr = "constant" )
Simple feature collection with 600 features and 0 fields
Geometry type: POINT
Dimension: XY
Bounding box: xmin: -5.138996 ymin: 41.36216 xmax: 9.361004 ymax: 50.86216
Geodetic CRS: WGS 84
First 10 features:
geometry
1 POINT (-5.138996 41.36216)
2 POINT (-4.638996 41.36216)
3 POINT (-4.138996 41.36216)
4 POINT (-3.638996 41.36216)
5 POINT (-3.138996 41.36216)
6 POINT (-2.638996 41.36216)
7 POINT (-2.138996 41.36216)
8 POINT (-1.638996 41.36216)
9 POINT (-1.138996 41.36216)
10 POINT (-0.6389965 41.36216)
Then, it can be converted to a sp object (needed because it is the expected input for krigin function):
<- as_Spatial (france_grid_sf)
Then, we can interpolate sunlight for each point of the grid. To that end, we use the krige() function from {gstat}. The varigram model is trained using the variogram() function from the same package.
Average over a long period
We create a function to perform the estimation for a specific month (we will therefore obtain climate data, as we will give as an input weather data from a long period).
#' Monthy sunlight average for a given month #' #' @param month month mois pour lequel realiser le krigeage #' month <- 1 <- function (month){# Keep only the month of interest <- |> ungroup () |> filter (month == !! month) |> filter (! is.na (INST))<- |> st_as_sf (coords = c ("Longitude" , "Latitude" ), crs = 4326 , agr = "constant" )<- |> as_Spatial ()# Variogramme <- gstat:: variogram (INST ~ 1 , df_ensoleillement_month_tmp_sp)# Select variogram model <- gstat:: fit.variogram (model = gstat:: vgm ("Gau" )# attr(variog_fr_fit, "singular") # Krigeage <- gstat:: krige (~ 1 ,model = variog_fr_fit<- |> tbl_df () |> rename (long = coords.x1, lat = coords.x2,ensol = var1.pred, ensol_var = var1.var|> mutate (mois = !! month)# End of monthly_sunshine_grid()
Let us loop over all month and apply this function to each of them:
<- map_df (1 : 12 , monthly_sunshine_grid)
[using ordinary kriging]
[using ordinary kriging]
[using ordinary kriging]
[using ordinary kriging]
[using ordinary kriging]
[using ordinary kriging]
[using ordinary kriging]
[using ordinary kriging]
[using ordinary kriging]
[using ordinary kriging]
[using ordinary kriging]
[using ordinary kriging]
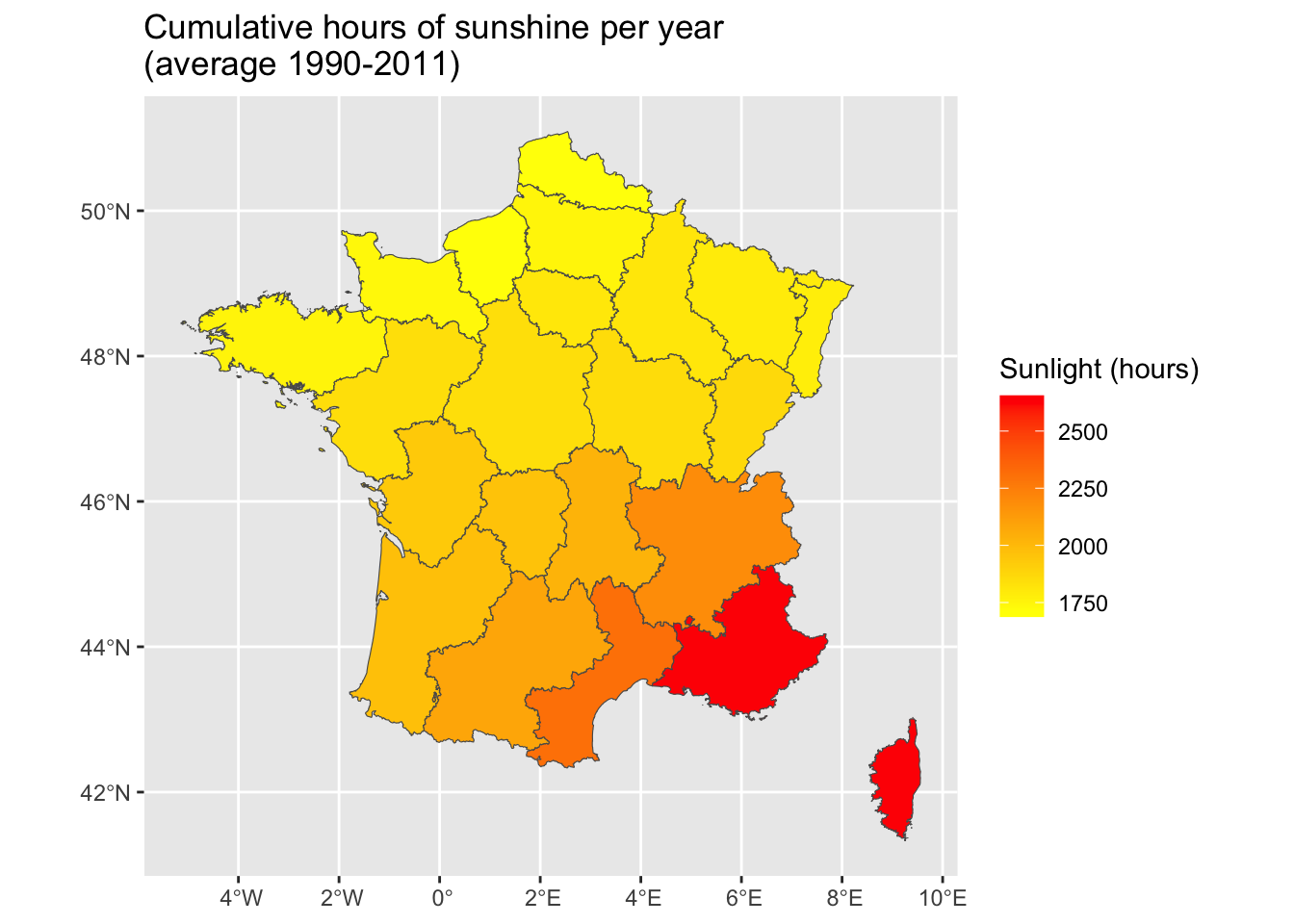
We can compute the average monthly sunlight over the period 1990-2011 as follows:
<- |> group_by (long, lat) |> # Cumulative hours summarise (ensol = sum (ensol)) |> ungroup () |> mutate (geometry = str_c ("POLYGON((" ,", " ,+ size_cell, lat, ", " ,+ size_cell, lat + size_cell, ", " ,+ size_cell,", " , "))" ,sep = " " |> st_as_sf (coords = c ("long" , "lat" ), wkt = "geometry" , crs = 4326 )
`summarise()` has grouped output by 'long'. You can override using the
`.groups` argument.
Simple feature collection with 600 features and 3 fields
Geometry type: POLYGON
Dimension: XY
Bounding box: xmin: -5.138996 ymin: 41.36216 xmax: 9.861004 ymax: 51.36216
Geodetic CRS: WGS 84
# A tibble: 600 × 4
long lat ensol geometry
* <dbl> <dbl> <dbl> <POLYGON [°]>
1 -5.14 41.4 1986. ((-5.138996 41.36216, -4.638996 41.36216, -4.638996 41.862…
2 -5.14 41.9 1989. ((-5.138996 41.86216, -4.638996 41.86216, -4.638996 42.362…
3 -5.14 42.4 1995. ((-5.138996 42.36216, -4.638996 42.36216, -4.638996 42.862…
4 -5.14 42.9 2001. ((-5.138996 42.86216, -4.638996 42.86216, -4.638996 43.362…
5 -5.14 43.4 2006. ((-5.138996 43.36216, -4.638996 43.36216, -4.638996 43.862…
6 -5.14 43.9 2007. ((-5.138996 43.86216, -4.638996 43.86216, -4.638996 44.362…
7 -5.14 44.4 2001. ((-5.138996 44.36216, -4.638996 44.36216, -4.638996 44.862…
8 -5.14 44.9 1987. ((-5.138996 44.86216, -4.638996 44.86216, -4.638996 45.362…
9 -5.14 45.4 1963. ((-5.138996 45.36216, -4.638996 45.36216, -4.638996 45.862…
10 -5.14 45.9 1927. ((-5.138996 45.86216, -4.638996 45.86216, -4.638996 46.362…
# ℹ 590 more rows
And then aggregate the values at the French region level:
<- |> :: st_join (map_departements, join = st_intersects, left = FALSE ) |> :: st_set_geometry (NULL ) |> select (CODE_DEPT, ensol) |> group_by (CODE_DEPT) |> summarise (ensol = mean (ensol))
# A tibble: 96 × 2
CODE_DEPT ensol
<chr> <dbl>
1 01 1967.
2 02 1779.
3 03 1898.
4 04 2688.
5 05 2542.
6 06 2753.
7 07 2299.
8 08 1786.
9 09 2094.
10 10 1840.
# ℹ 86 more rows
Let us visualize the results:
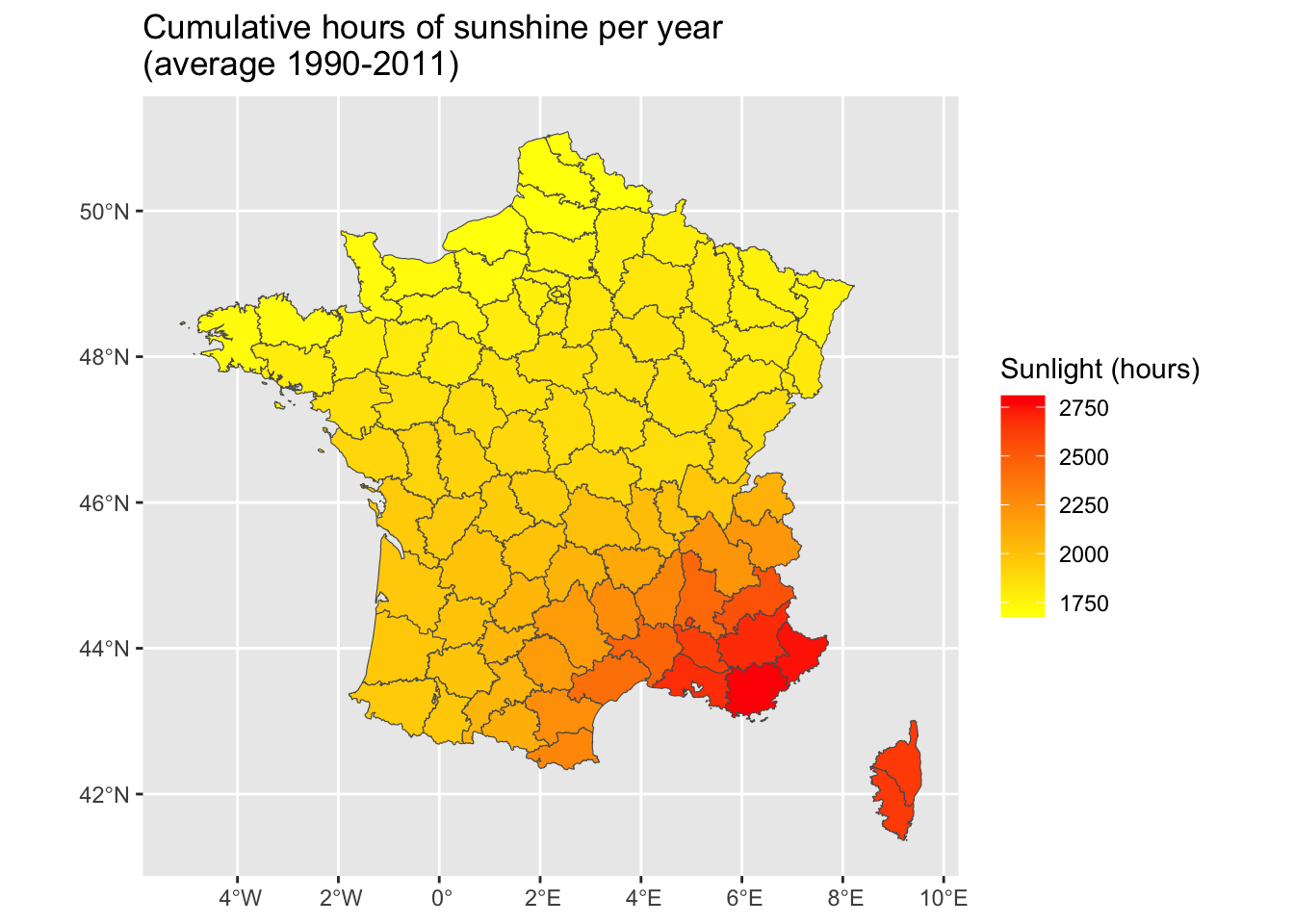
<- ggplot (data = map_departements |> left_join (ensol_regions)) + geom_sf (mapping = aes (fill = ensol)) + scale_fill_gradient ("Sunlight (hours)" , low= "yellow" , high= "red" ) + labs (title = "Cumulative hours of sunshine per year \n (average 1990-2011)" )
Joining with `by = join_by(CODE_DEPT)`
Sunlight for 2011
Let us look at the cumulative value for 2011:
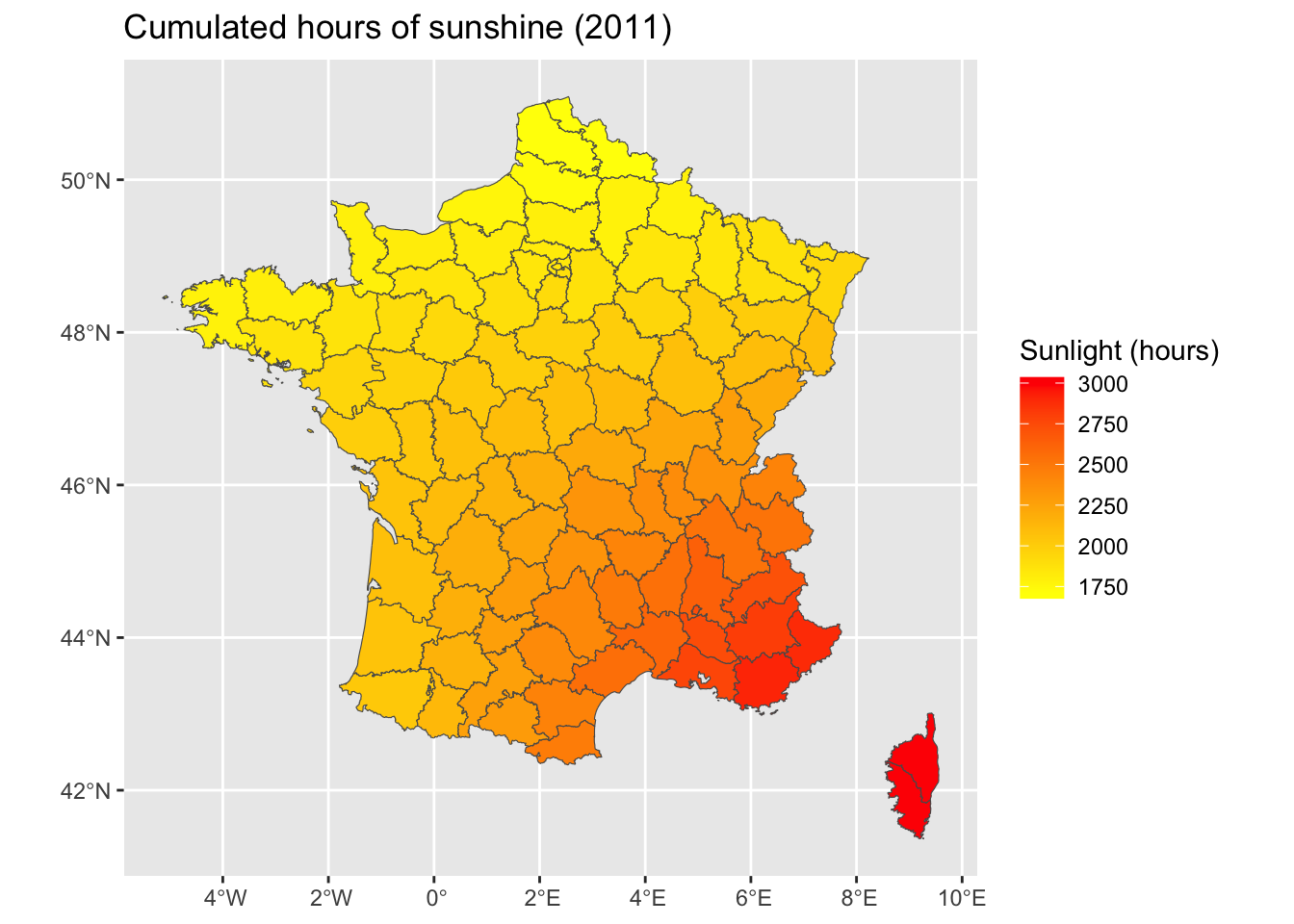
<- |> mutate (month = lubridate:: month (DAT), year = lubridate:: year (DAT)) |> filter (year == 2011 ) |> filter (! is.na (INST)) |> group_by (NUM_POSTE) |> mutate (n = n ()) |> filter (n == 12 ) |> group_by (NUM_POSTE) |> summarise (cumul_ensol = sum (INST, na.rm= TRUE )) |> left_join (stations, by = c ("NUM_POSTE" = "ID" ))
# A tibble: 41 × 6
NUM_POSTE cumul_ensol Nom Latitude Longitude Altitude
<chr> <dbl> <chr> <dbl> <dbl> <dbl>
1 07005 1591 ABBEVILLE 50.1 1.83 69
2 07015 1705 LILLE-LESQUIN 50.6 3.10 47
3 07027 1905 CAEN-CARPIQUET 49.2 -0.456 67
4 07037 1611 ROUEN-BOOS 49.4 1.18 151
5 07110 1610 BREST-GUIPAVAS 48.4 -4.41 94
6 07130 1935 RENNES-ST JACQUES 48.1 -1.73 36
7 07139 1821 ALENCON 48.4 0.110 143
8 07149 1882 ORLY 48.7 2.38 89
9 07168 2014 TROYES-BARBEREY 48.3 4.02 112
10 07190 1898 STRASBOURG-ENTZHEIM 48.5 7.64 150
# ℹ 31 more rows
Filtering the results to keep only stations in Metropolitan France:
<- |> filter (>= bbox_france["xmin" ],<= bbox_france["xmax" ],>= bbox_france["ymin" ],<= bbox_france["ymax" ]
The coordinates of weather stations:
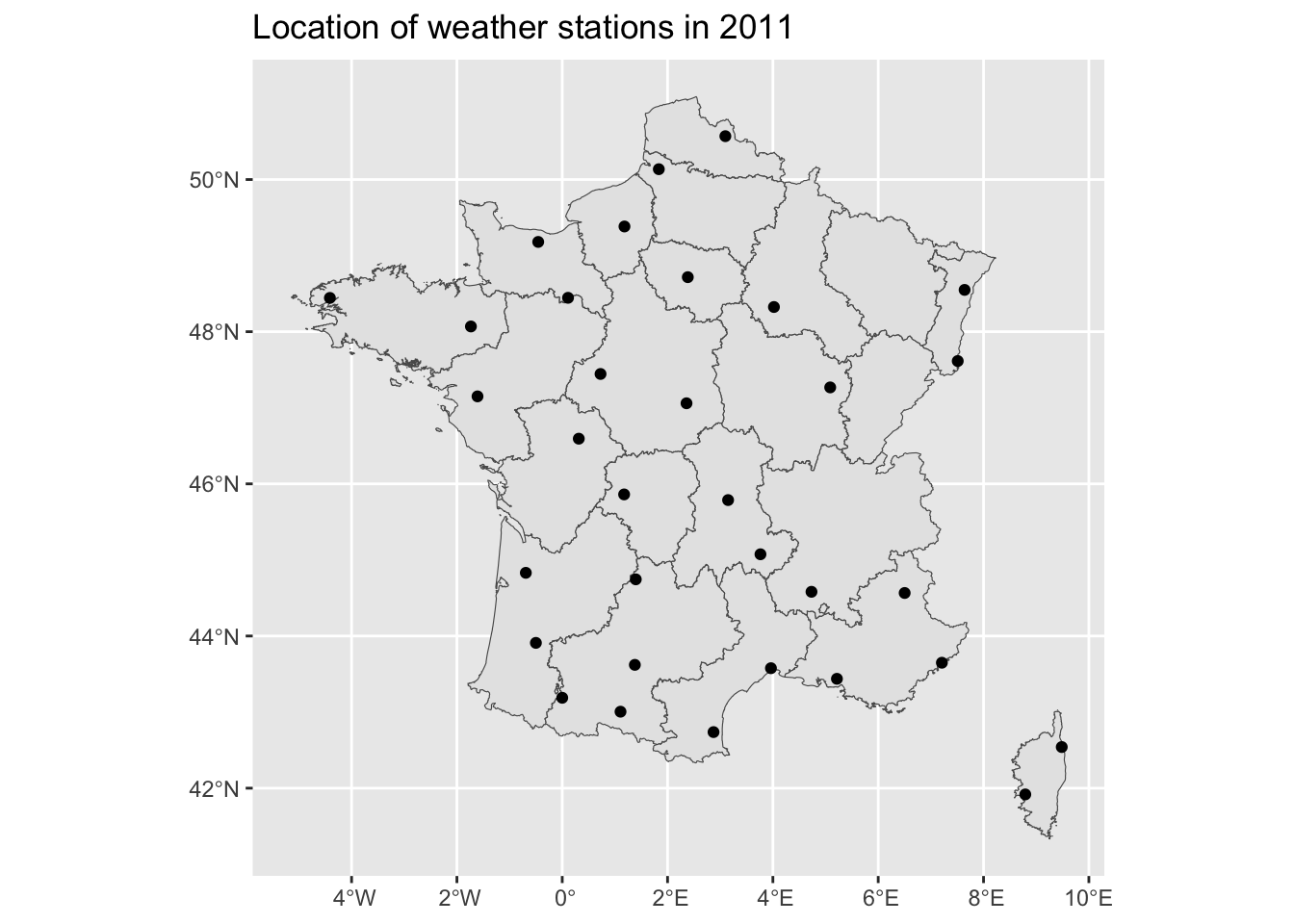
<- |> select ("Longitude" , "Latitude" ) |> unique () |> st_as_sf (coords = c ("Longitude" , "Latitude" )) |> :: st_set_crs (4326 )
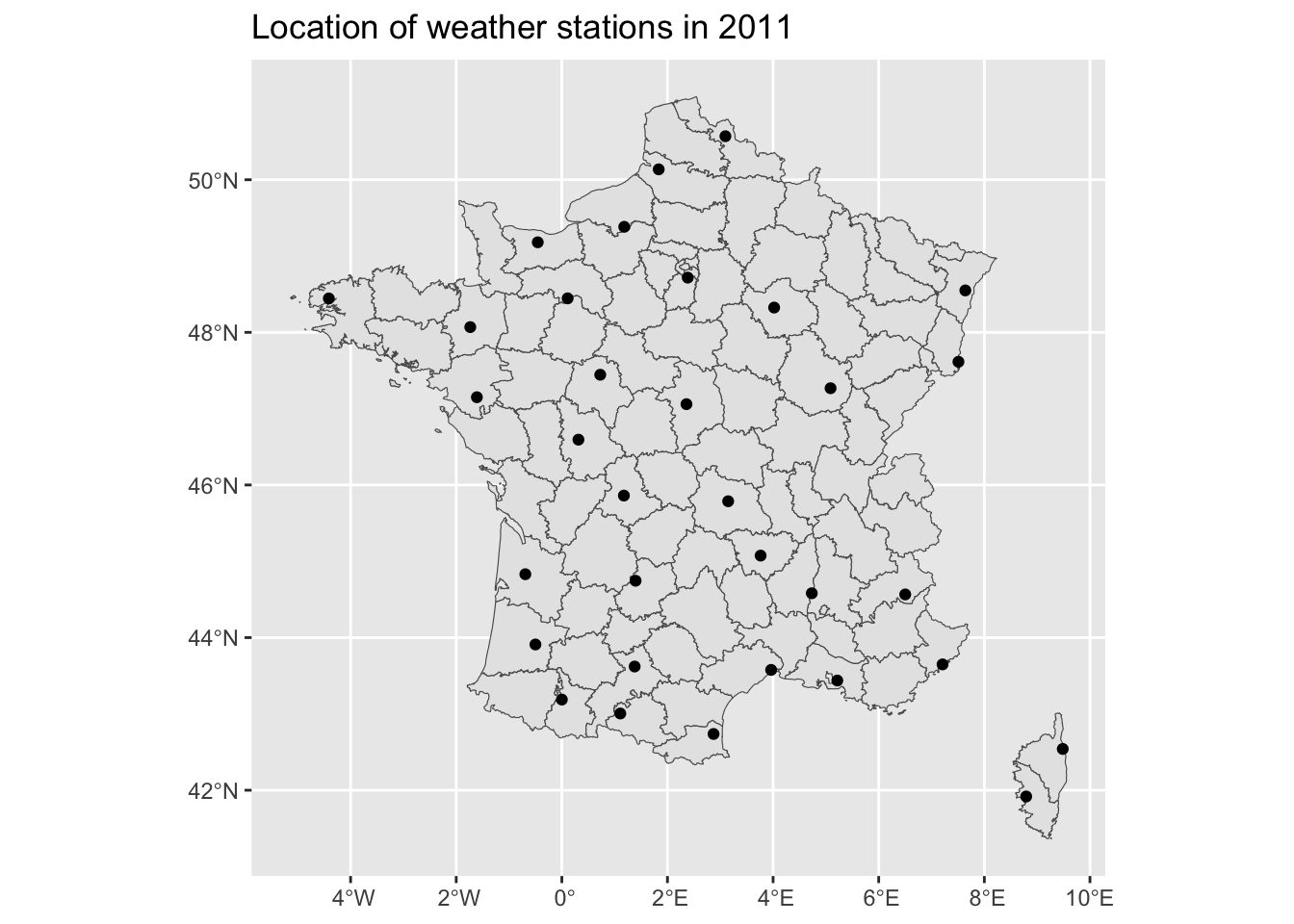
And a graph of where those stations are located:
<- ggplot () + geom_sf (data = map_departements) + scale_fill_discrete (guide = FALSE ) + geom_sf (data = coords_stations_2011) + labs (title = "Location of weather stations in 2011" )
Warning: The `guide` argument in `scale_*()` cannot be `FALSE`. This was deprecated in
ggplot2 3.3.4.
ℹ Please use "none" instead.
Preparation of the sp object for the grid covering France:
<- |> st_as_sf (coords = c ("long" , "lat" ), crs = 4326 , agr = "constant" )<- as_Spatial (france_grid_sf)
Let us turn df_ensoleillement_2011 to an sf object, and then as an sp one:
<- |> st_as_sf (coords = c ("Longitude" , "Latitude" ), crs = 4326 , agr = "constant" )<- |> as_Spatial ()
Now let us turn to the interpolation. First, let us fit a variogram model to the data:
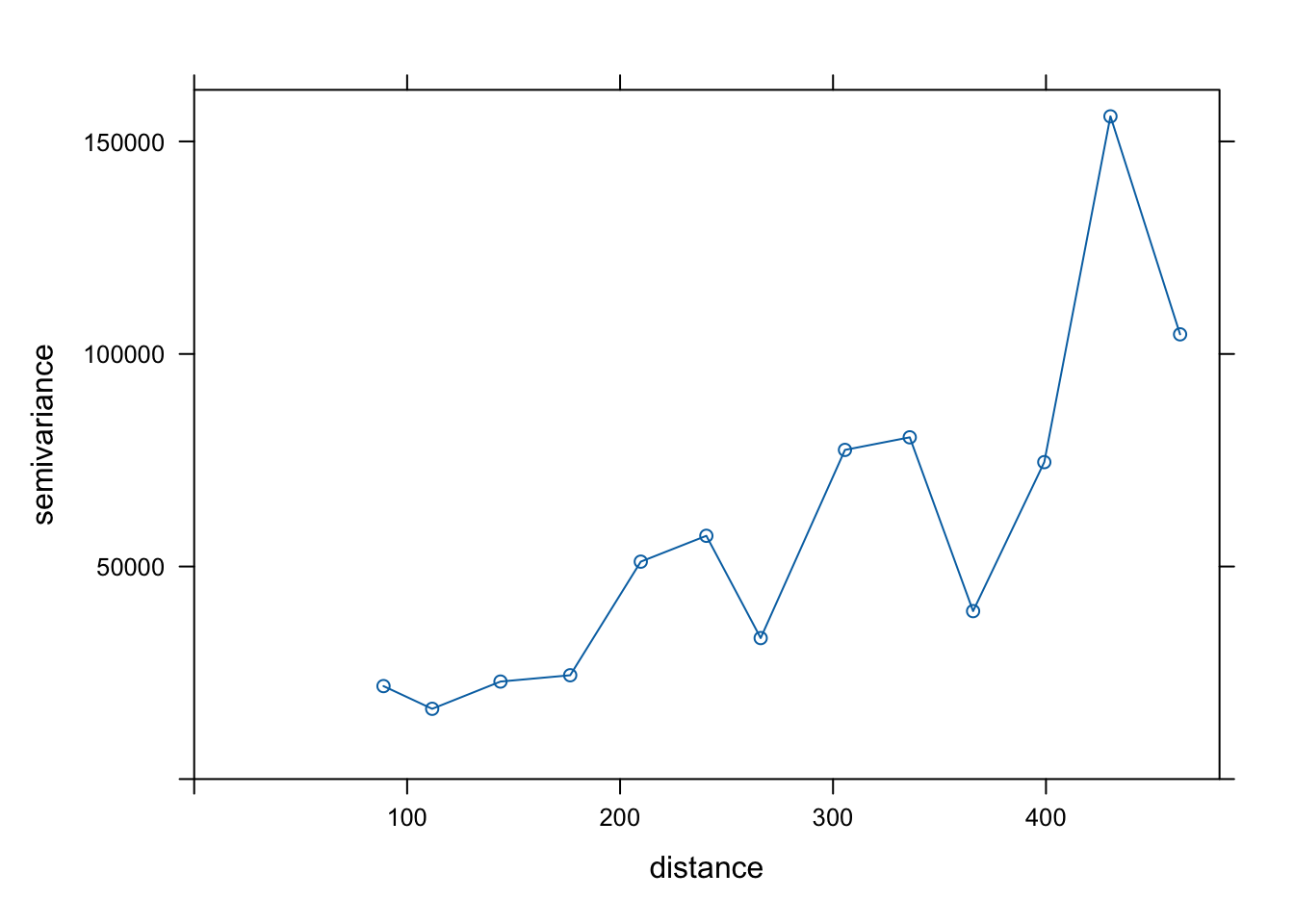
<- :: variogram (cumul_ensol ~ 1 , df_ensoleillement_2011_sp)
Let us select a variogram model:
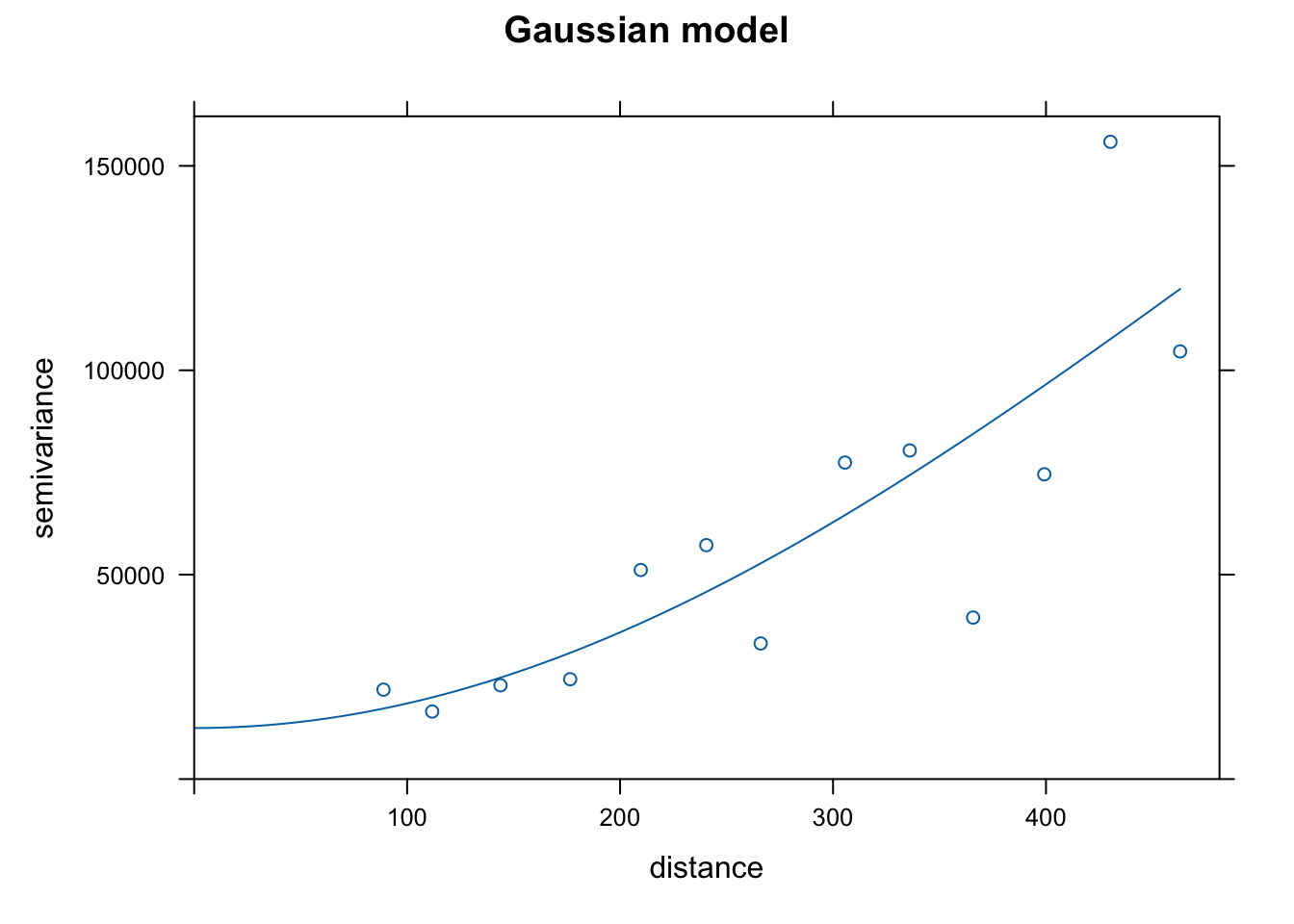
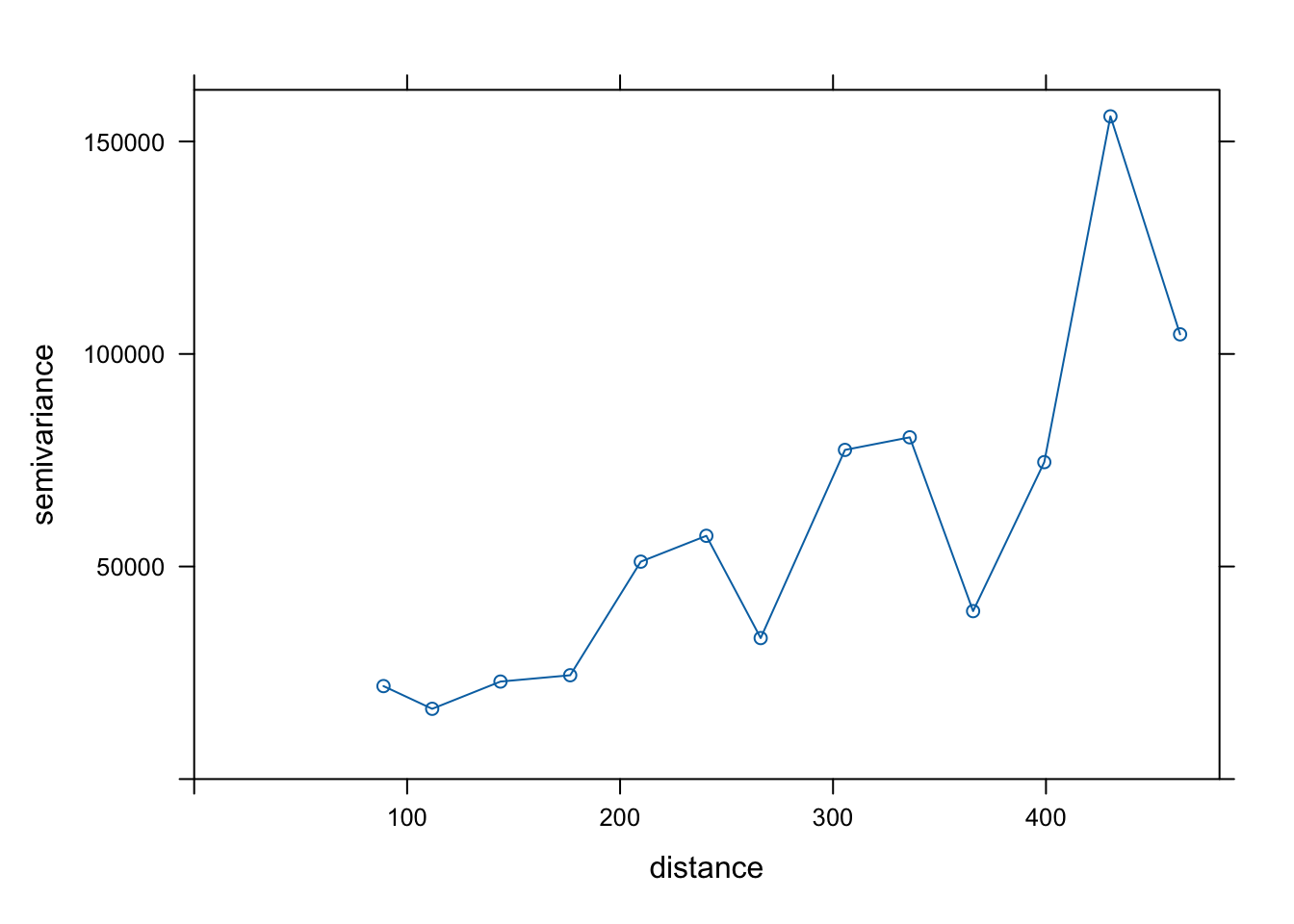
<- gstat:: fit.variogram (variog_fr, model = gstat:: vgm ("Gau" ))
Checking whether the algorithm converged:
attr (variog_fr_fit, "singular" )
Quick graph of the fit:
plot (variog_fr, variog_fr_fit, main = 'Gaussian model' )
Now let us interpolate the data at each point of the grid:
<- :: krige (cumul_ensol ~ 1 ,model= variog_fr_fit)
Let us turn the results into a tibble:
<- |> tbl_df () |> rename (long = coords.x1, lat = coords.x2,ensol = var1.pred, ensol_var = var1.var)
Warning: `tbl_df()` was deprecated in dplyr 1.0.0.
ℹ Please use `tibble::as_tibble()` instead.
# A tibble: 600 × 4
long lat ensol ensol_var
<dbl> <dbl> <dbl> <dbl>
1 -5.14 41.4 1721. 141302.
2 -4.64 41.4 1726. 123759.
3 -4.14 41.4 1736. 107596.
4 -3.64 41.4 1752. 92994.
5 -3.14 41.4 1774. 80068.
6 -2.64 41.4 1803. 68871.
7 -2.14 41.4 1840. 59390.
8 -1.64 41.4 1885. 51553.
9 -1.14 41.4 1938. 45242.
10 -0.639 41.4 1999. 40306.
# ℹ 590 more rows
Let us create an sf object from these results:
<- |> group_by (long, lat) |> # Cumulative hours summarise (ensol = sum (ensol)) |> ungroup () |> mutate (geometry = str_c ("POLYGON((" ,", " ,+ size_cell, lat, ", " ,+ size_cell, lat + size_cell, ", " ,+ size_cell,", " , "))" ,sep = " " |> st_as_sf (coords = c ("long" , "lat" ), wkt = "geometry" ,crs = 4326 )
`summarise()` has grouped output by 'long'. You can override using the
`.groups` argument.
And then we can get the cumulative sunlight over the year, by French département:
<- |> :: st_join (map_departements, join = st_intersects, left = FALSE ) |> :: st_set_geometry (NULL ) |> select (CODE_DEPT, ensol) |> group_by (CODE_DEPT) |> summarise (ensol = mean (ensol))
And now the results can be graphed:
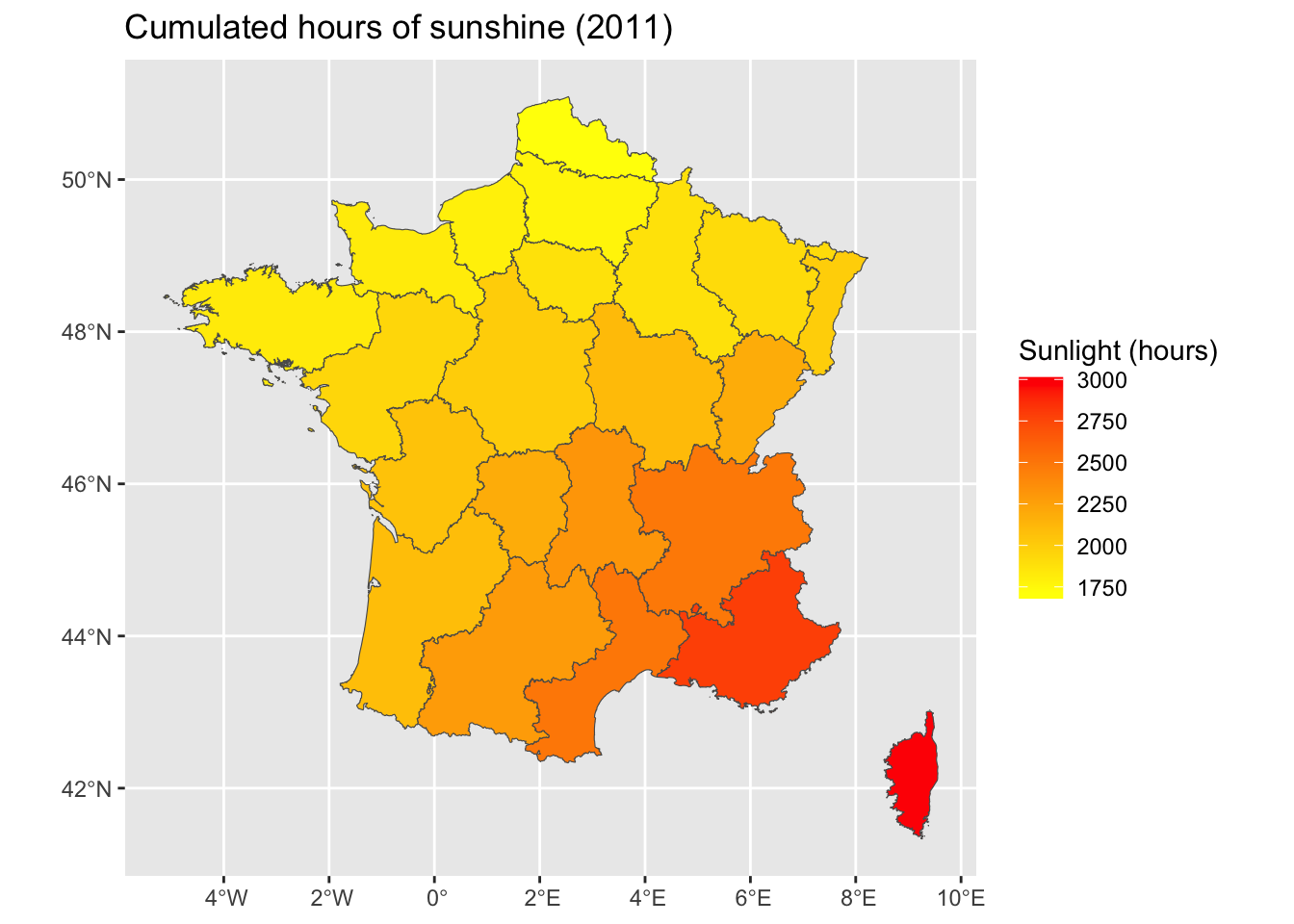
<- ggplot (data = map_departements |> left_join (ensol_regions_2011)) + geom_sf (aes (fill = ensol)) + scale_fill_gradient ("Sunlight (hours)" , low= "yellow" , high= "red" ) + labs (title = "Cumulated hours of sunshine (2011)" )
Joining with `by = join_by(CODE_DEPT)`
At the level of French “régions”
Now let us interpolate sunlight over the French Regions (with their geographical boundaries as of 2014).
load ("../data/map_regions_2014.rda" )
The bounding box:
<- st_bbox (map_regions_2014)
old-style crs object detected; please recreate object with a recent sf::st_crs()
Keeping only weather stations from Metropolitan France:
<- |> filter (Longitude >= bbox_france["xmin" ],<= bbox_france["xmax" ],>= bbox_france["ymin" ],<= bbox_france["ymax" ])
Coordinates of those stations:
<- |> select ("Longitude" , "Latitude" ) |> unique () |> st_as_sf (coords = c ("Longitude" , "Latitude" )) |> :: st_set_crs (4326 )
Adding missing grouping variables: `NUM_POSTE`
Simple feature collection with 40 features and 1 field
Geometry type: POINT
Dimension: XY
Bounding box: xmin: -4.412 ymin: 41.918 xmax: 9.485167 ymax: 50.57
Geodetic CRS: WGS 84
# A tibble: 40 × 2
# Groups: NUM_POSTE [40]
NUM_POSTE geometry
* <chr> <POINT [°]>
1 07005 (1.834 50.136)
2 07015 (3.0975 50.57)
3 07020 (-1.939833 49.72517)
4 07027 (-0.456167 49.18)
5 07037 (1.181667 49.383)
6 07110 (-4.412 48.44417)
7 07117 (-3.473167 48.82583)
8 07130 (-1.734 48.06883)
9 07139 (0.110167 48.4455)
10 07149 (2.384333 48.71683)
# ℹ 30 more rows
The kriging wraping functions:
#' Average Monthly sunlight for a given month (interpolated with kriging methods) #' at the French Region level #' @param month month to select for kriging #' month <- 1 <- function (month){# Keep only the month of interest <- |> ungroup () |> filter (month == !! month) |> filter (! is.na (INST))<- |> st_as_sf (coords = c ("Longitude" , "Latitude" ), crs = 4326 , agr = "constant" )<- |> as_Spatial ()# Variogram <- :: variogram (INST ~ 1 , df_ensoleillement_month_tmp_sp)# Select variogram model <- gstat:: fit.variogram (model = gstat:: vgm ("Gau" )attr (variog_fr_fit, "singular" )# Kriging <- gstat:: krige (INST ~ 1 ,model= variog_fr_fit)<- |> tbl_df () |> rename (long = coords.x1, lat = coords.x2,ensol = var1.pred, ensol_var = var1.var) |> mutate (mois = !! month)# End of monthly_sunshine_grid()
Looping over the months:
<- map_df (1 : 12 , monthly_sunshine_grid)
[using ordinary kriging]
[using ordinary kriging]
[using ordinary kriging]
[using ordinary kriging]
[using ordinary kriging]
[using ordinary kriging]
[using ordinary kriging]
[using ordinary kriging]
[using ordinary kriging]
[using ordinary kriging]
[using ordinary kriging]
[using ordinary kriging]
Average over a long period
Annual average of cumulative over the 1990-2011 period:
<- |> group_by (long, lat) |> # Cumulative hours summarise (ensol = sum (ensol)) |> ungroup () |> mutate (geometry = str_c ("POLYGON((" ,", " ,+ size_cell, lat, ", " ,+ size_cell, lat + size_cell, ", " ,+ size_cell,", " , "))" ,sep = " " |> st_as_sf (coords = c ("long" , "lat" ), wkt = "geometry" ,crs = 4326 )
`summarise()` has grouped output by 'long'. You can override using the
`.groups` argument.
Values on a tibble, each line representing a French Region:
<- |> :: st_join (map_regions_2014, join = st_intersects, left = FALSE ) |> :: st_set_geometry (NULL ) |> select (code_insee, ensol) |> group_by (code_insee) |> summarise (ensol = mean (ensol))
old-style crs object detected; please recreate object with a recent sf::st_crs()
old-style crs object detected; please recreate object with a recent sf::st_crs()
old-style crs object detected; please recreate object with a recent sf::st_crs()
old-style crs object detected; please recreate object with a recent sf::st_crs()
# A tibble: 22 × 2
code_insee ensol
<chr> <dbl>
1 11 1806.
2 21 1809.
3 22 1745.
4 23 1708.
5 24 1844.
6 25 1734.
7 26 1851.
8 31 1688.
9 41 1787.
10 42 1777.
# ℹ 12 more rows
Visualize on a map:
<- ggplot (data = map_regions_2014 |> left_join (ensol_regions)) + geom_sf (aes (fill = ensol)) + scale_fill_gradient ("Sunlight (hours)" , low= "yellow" , high= "red" ) + labs (title = "Cumulative hours of sunshine per year \n (average 1990-2011)" )
Joining with `by = join_by(code_insee)`
old-style crs object detected; please recreate object with a recent
sf::st_crs()
Sunlight for 2011
<- |> mutate (month = lubridate:: month (DAT), year = lubridate:: year (DAT)) |> filter (year == 2011 ) |> filter (! is.na (INST)) |> group_by (NUM_POSTE) |> mutate (n = n ()) |> filter (n == 12 ) |> group_by (NUM_POSTE) |> summarise (cumul_ensol = sum (INST, na.rm= TRUE )) |> left_join (stations, by = c ("NUM_POSTE" = "ID" ))
# A tibble: 41 × 6
NUM_POSTE cumul_ensol Nom Latitude Longitude Altitude
<chr> <dbl> <chr> <dbl> <dbl> <dbl>
1 07005 1591 ABBEVILLE 50.1 1.83 69
2 07015 1705 LILLE-LESQUIN 50.6 3.10 47
3 07027 1905 CAEN-CARPIQUET 49.2 -0.456 67
4 07037 1611 ROUEN-BOOS 49.4 1.18 151
5 07110 1610 BREST-GUIPAVAS 48.4 -4.41 94
6 07130 1935 RENNES-ST JACQUES 48.1 -1.73 36
7 07139 1821 ALENCON 48.4 0.110 143
8 07149 1882 ORLY 48.7 2.38 89
9 07168 2014 TROYES-BARBEREY 48.3 4.02 112
10 07190 1898 STRASBOURG-ENTZHEIM 48.5 7.64 150
# ℹ 31 more rows
Metropolitan French stations only:
<- |> filter (Longitude >= bbox_france["xmin" ],<= bbox_france["xmax" ],>= bbox_france["ymin" ],<= bbox_france["ymax" ])
Their coordinates:
<- |> select ("Longitude" , "Latitude" ) |> unique () |> st_as_sf (coords = c ("Longitude" , "Latitude" )) |> :: st_set_crs (4326 )
Visualize their locations:
<- ggplot () + geom_sf (data = map_regions_2014) + scale_fill_discrete (guide = FALSE ) + geom_sf (data = coords_stations_2011) + labs (title = "Location of weather stations in 2011" )
old-style crs object detected; please recreate object with a recent sf::st_crs()
old-style crs object detected; please recreate object with a recent sf::st_crs()
Preparing the sp object for kriging:
<- |> st_as_sf (coords = c ("Longitude" , "Latitude" ), crs = 4326 , agr = "constant" )
<- |> as_Spatial ()
The variogram model can be fitted:
<- :: variogram (cumul_ensol ~ 1 , df_ensoleillement_2011_sp)
Plotting the results:
Selection of the best fit:
<- gstat:: fit.variogram (variog_fr, model = gstat:: vgm ("Gau" ))
Checking whether the algorithm converged:
attr (variog_fr_fit, "singular" )
Kriging:
<- gstat:: krige (~ 1 ,model= variog_fr_fit
The interpolated values can be stored on a tibble:
<- |> tbl_df () |> rename (long = coords.x1, lat = coords.x2,ensol = var1.pred, ensol_var = var1.var
Warning: `tbl_df()` was deprecated in dplyr 1.0.0.
ℹ Please use `tibble::as_tibble()` instead.
# A tibble: 600 × 4
long lat ensol ensol_var
<dbl> <dbl> <dbl> <dbl>
1 -5.14 41.4 1721. 141302.
2 -4.64 41.4 1726. 123759.
3 -4.14 41.4 1736. 107596.
4 -3.64 41.4 1752. 92994.
5 -3.14 41.4 1774. 80068.
6 -2.64 41.4 1803. 68871.
7 -2.14 41.4 1840. 59390.
8 -1.64 41.4 1885. 51553.
9 -1.14 41.4 1938. 45242.
10 -0.639 41.4 1999. 40306.
# ℹ 590 more rows
And those can be put on an sf object:
<- |> group_by (long, lat) |> # Cumul heures summarise (ensol = sum (ensol)) |> ungroup () |> mutate (geometry = str_c ("POLYGON((" ,", " ,+ size_cell, lat, ", " ,+ size_cell, lat + size_cell, ", " ,+ size_cell,", " , "))" ,sep = " " |> st_as_sf (coords = c ("long" , "lat" ), wkt = "geometry" ,crs = 4326 )
`summarise()` has grouped output by 'long'. You can override using the
`.groups` argument.
The average value at the region level, for 2011:
<- |> :: st_join (map_regions_2014, join = st_intersects, left = FALSE ) |> :: st_set_geometry (NULL ) |> select (code_insee, ensol) |> group_by (code_insee) |> summarise (ensol = mean (ensol))
old-style crs object detected; please recreate object with a recent sf::st_crs()
old-style crs object detected; please recreate object with a recent sf::st_crs()
old-style crs object detected; please recreate object with a recent sf::st_crs()
old-style crs object detected; please recreate object with a recent sf::st_crs()
# A tibble: 22 × 2
code_insee ensol
<chr> <dbl>
1 11 1876.
2 21 1880.
3 22 1771.
4 23 1790.
5 24 2004.
6 25 1819.
7 26 2102.
8 31 1681.
9 41 1915.
10 42 1991.
# ℹ 12 more rows
Choropleth map of these values:
<- ggplot (data = map_regions_2014 |> left_join (ensol_regions_2011)) + geom_sf (aes (fill = ensol)) + scale_fill_gradient ("Sunlight (hours)" , low= "yellow" , high= "red" ) + labs (title = "Cumulated hours of sunshine (2011)" )
Joining with `by = join_by(code_insee)`
old-style crs object detected; please recreate object with a recent
sf::st_crs()
Lastly, these results can be saved into an RData object:
<- |> rename (ensol_1990_2011 = ensol) |> left_join (ensol_regions_2011 |> rename (ensol_2011 = ensol))save (ensol_regions, file = "../data/meteo/ensol_regions.rda" )